On this page...
Overview
The 2000s saw an explosion of population-scale genome-wide association studies (GWAS). Seeing a need to accelerate and coordinate post-GWAS research to advance our understanding of the genetic architecture of cancer, NCI spearheaded the GAME-ON initiative.
This initiative, focused on five cancer types, brought together genetic epidemiologists and biologists, along with other disciplines, to build a cross-discipline collaboration to empower rapid replication of GWAS results and additional discovery, particularly in understudied populations.
The five cancer sites studied in the GAME-ON initiative are listed below. Descriptions of each project, and their publications, are available through the link in each bullet to NIH’s Research Portfolio Online Reporting Tools (NIH RePORTer).
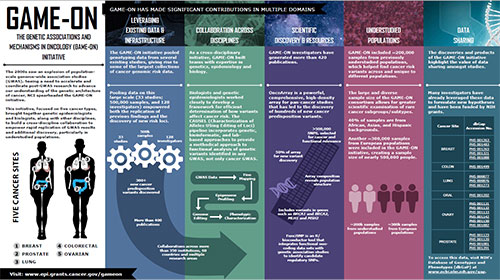
Infographic
Learn more about the Genetic Associations and Mechanisms in Oncology (GAME-ON) initiative.
View the Infographic [PDF - 480 KB]Leveraging Existing Data and Infrastructure
The GAME-ON initiative pooled genotyping data from several existing studies, giving rise to some of the largest collections of cancer genomic risk data.
Pooling data on this large scale (33 studies; 500,000 samples, and 128 investigators) empowered both the replication of previous findings and the discovery of new risk loci.
Collaboration Across Disciplines
As a cross-disciplinary initiative, GAME-ON built teams with expertise in genetics, epidemiology and biology. GAME-ON included collaborations across more than 350 institutions, 60 countries, and multiple research areas.
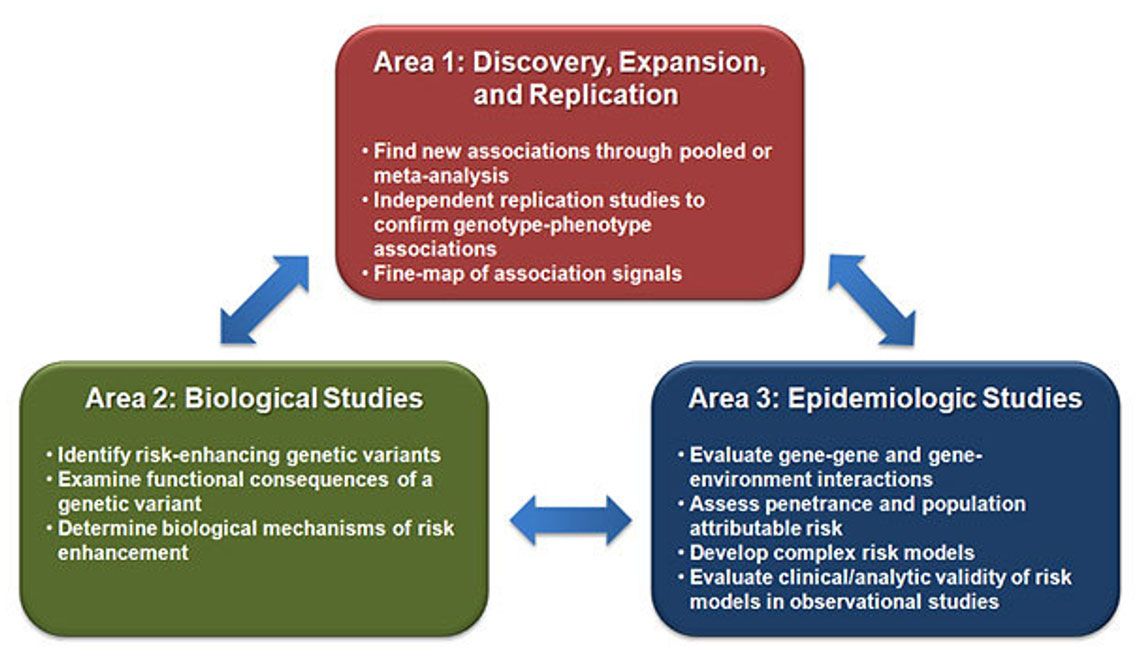
Area 1: Discovery, Expansion, and Replication. Area 2: Biological Studies. Area 3: Epidemiologic Studies.
Biologists and genetic epidemiologists worked closely to develop a framework for efficient determination of how variants affect cancer risk. The CAUSEL (Characterization of Alleles USing Editing of Loci) pipeline incorporates genetic, bioinformatic, and lab-based approaches to provide a methodical approach to functional analysis of genetic variants identified in any GWAS, not only cancer GWAS.
GWAS Data ![]() Fine Mapping
Fine Mapping ![]() Epigenome Profiling
Epigenome Profiling ![]() Genome Editing
Genome Editing ![]() Phenotypic Characterization
Phenotypic Characterization
Scientific Discovery and Resources
GAME-ON investigators have generated more than 420 publications. GAME-ON researchers also developed datasets, tools and efficiencies that aided discovery within this initiative and beyond.
For questions related to the GAME-ON grant awards:

Stefanie Nelson, PhD
Program Director, Genomic Epidemiology Branch
nelsons@mail.nih.gov
- A variety of datasets associated with this initiative have been released for continued scientific use.
- OncoArray is a powerful, comprehensive, high-density array for pan-cancer studies that has led to the discovery of hundreds of new cancer predisposition variants.
Features of the OncoArray include:- >500,000 single nucleotide polymorphisms (SNPs), selected for cancer and functional relevance
- 50% of array for new variant discovery
- Array composition reveals population structure
- Includes variants in key cancer genes, such as BRCA1 and BRCA2, MLH1 and MHS2
OncoArray was funded through substantial grants from NCI through the GAME-ON initiative and the Division of Cancer Epidemiology and Genetics (DCEG), Genome Canada/Génome Québec/Canadian Institutes of Health Research (CIHR) through the Personalised Risk Stratification for Prevention and Early Detection of Breast Cancer international project, Cancer Research UK (University of Cambridge), and a grant from the European Union's Seventh Framework Programme for Research (FP7) for the Collaborative Oncological Gene-environment Study (COGS), together with many other smaller grants.
- FunciSNP is an R/bioconductor
 tool that integrates functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs.
tool that integrates functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs.
Understudied Populations
GAME-ON included ~200,000 samples from previously understudied populations, which helped find cancer risk variants across and unique to different populations. 40% of samples are from African, Asian, and Hispanic backgrounds. Another ~300,000 samples from European populations were included in the GAME-ON initiative, creating a total sample size of nearly 500,000 people. The large and diverse sample size of GAME-ON allows for greater scientific examination of rare subgroups and subtypes.
Data Sharing
The discoveries and products of the GAME-ON initiative highlight the value of data sharing amongst studies. Below are links to data shared through NIH’s database of Genotypes and Phenotypes (dbGaP).
- Breast
- Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) – Genome-Wide Association Study Meta-Analysis
- DRIVE – germline whole genome sequencing
- DRIVE – OncoArray Genotypes
- Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) – OncoArray Genotypes
- Shanghai Breast Cancer Genetics Study
- Lung
- Ovarian
- Prostate
Many investigators have already leveraged these data to formulate new hypotheses and have been funded by NIH grants.